A Measure for Reliable Modelability of Protein Tertiary Structures
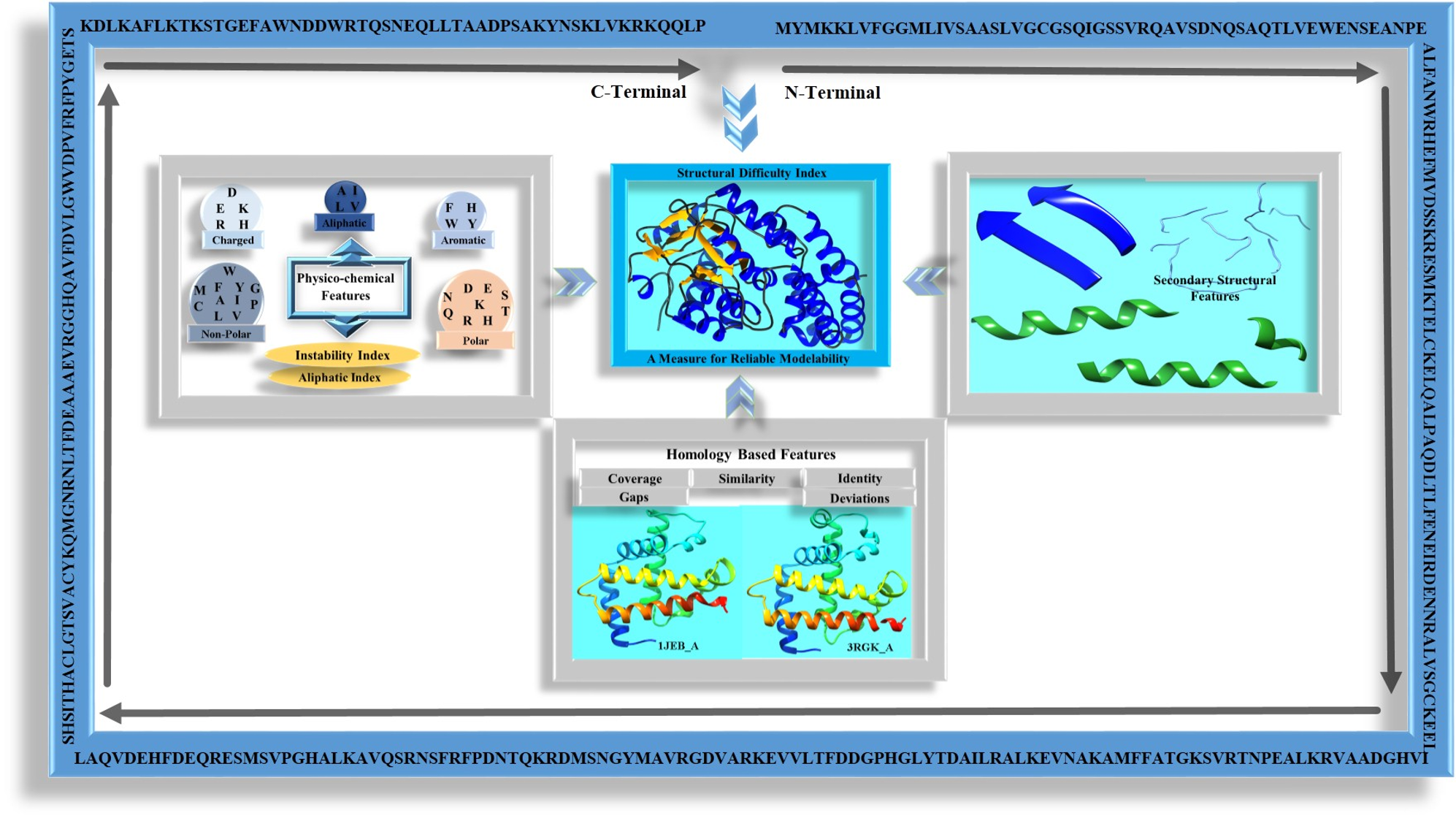
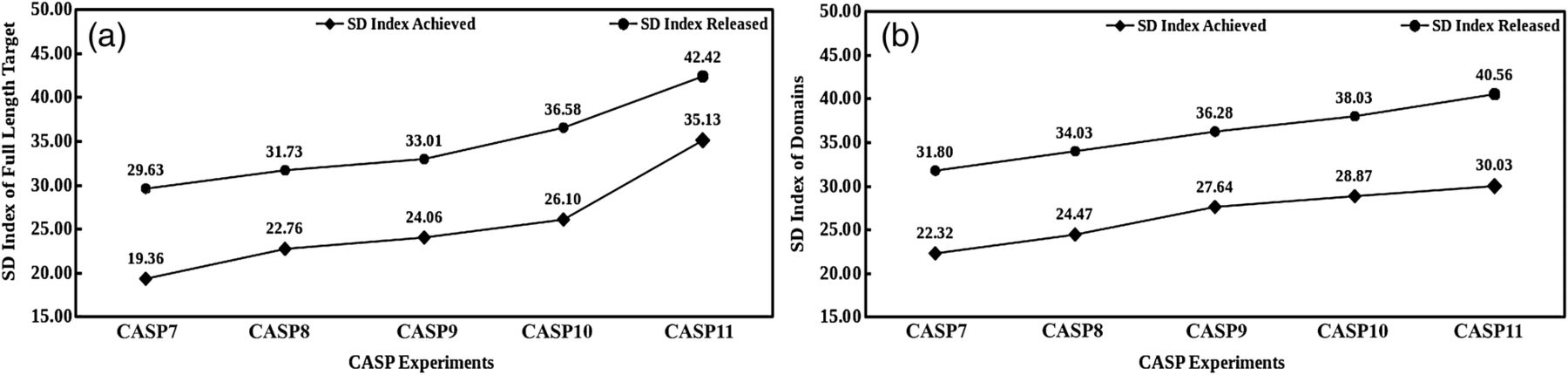
Where is the protein structure prediction field today or what is the success rate of best modeling softwares /servers for protein structure prediction ? Simple questions like these do not have simple and ready answers today. Conventional methods of explaining whether structure of a sequence is modelable in terms of homology based parameters do not reflect the current status of the field. There is more to structure prediction than mere homology. Similarity and identity based measures of modelability of a protein sequence may be misleading sometimes. “Structural Difficulty (SD)” index, which is derived from secondary structures, homology and physico-chemical features of protein sequences, reflects the capability of predicting good quality structures with some of the best methodologies available currently. The SD index also helps to assess the plausiblity for developing proteome level structural databases for various organisms.
In concordance with the present status and the rate of developments of proteins structure prediction field, the SD index scores can be categorized into three major classes, viz. modelable zone (represents present status), difficult zone (expected to be covered post-CASP11 with current rate of improvements) and very difficult zone (further improvements required post-CASP11 developments). The modelable zone accounts for the SD index under a score of 35 for a novel protein sequence and 30 for a protein sequence with known domain information. For the protein scoring in the modelable zone, a good quality (up to 3 Å rmsd from native) protein structure can be predicted with the present methods. The difficult zone addresses the sequences having SD index scores ranging from 35 to 50 where medium-quality structures may be modeled and which is likely to be pushed into modelable zone with recent improvements. The very difficult zone accounts for protein sequences having SD index scores above 50. These sequences are very difficult to model with present methodologies.