 |
Validation of Bhageerath |
| PDB ID | 1DFN | | Protein Name | Crystal Structure of Defensin HNP-3, an Amphiphilic Dimer Mechanisms of Membrane Permeabilization | | Sequence (FASTA format) | DCYCRIPACIAGERRYGTCIYQGRLWAFCC | | Secondary Structure Information | S 3-5, S 14-21, S 24-30 | | Native Structure | 1dfn |
| H - Helix, B - Beta strand |
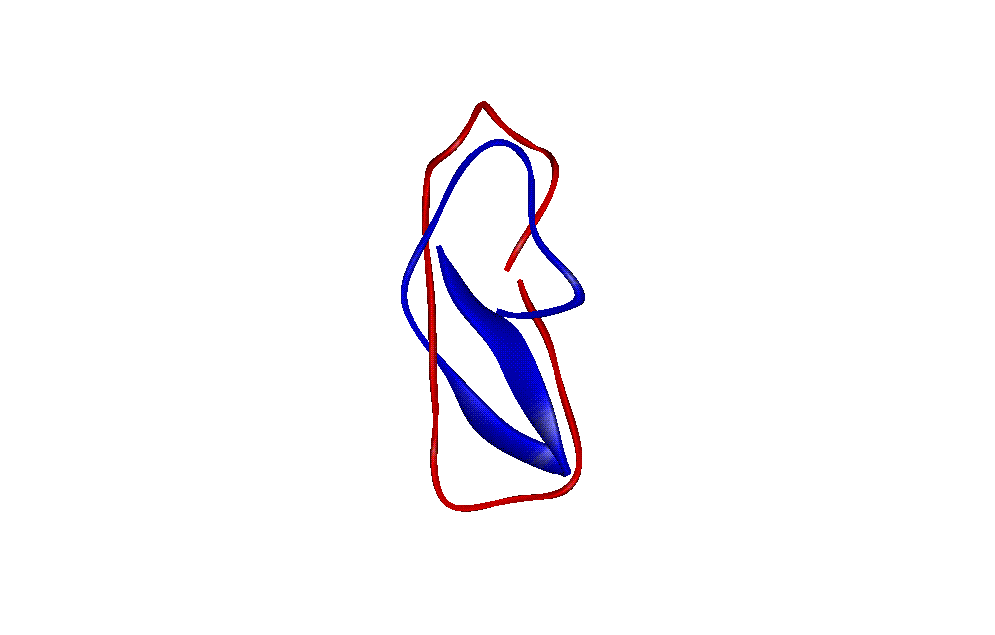 | The figure shows the Superimposition of the native structure with the lowest RMSD structure (PDB1) from the 10 lowest energy structures (Native is shown in Blue and lowest RMSD structure is in Red) |
|
|
|
© Copyright 2004-2009, Prof B. Jayaram & Co-workers| Disclaimer |
|
|
|
|

