RM2TS: From Ramchandran Maps to
Tertiary Structures of Proteins.
The missing link between Ramachandran map and tertiary structure

The missing link between Ramachandran map and tertiary structure
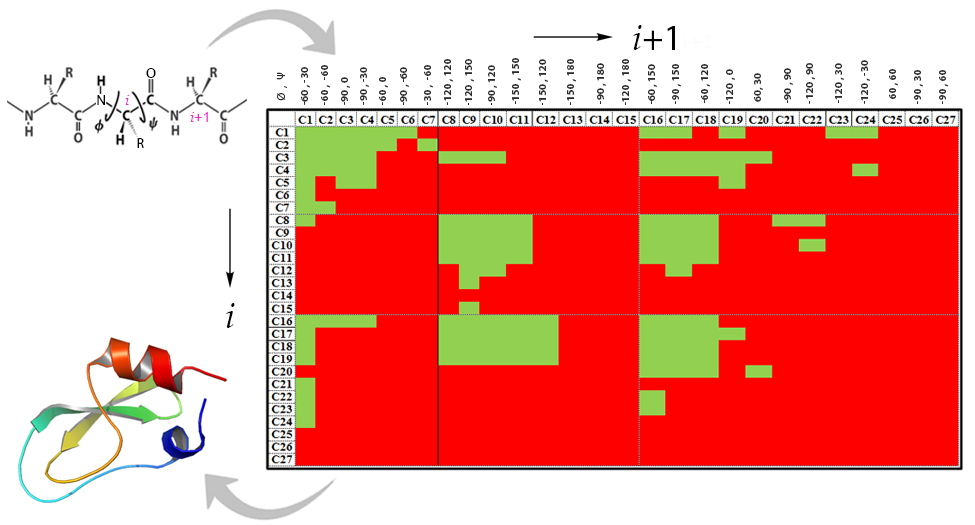
We have divided the allowed (Φ, Ψ) space in Ramachandran maps into 27 distinct conformations sufficient to regenerate a structure to within 5 Å from the native, at least for small proteins, thus reducing the structure prediction problem to a specification of an alphanumeric string i.e. amino acid sequence together with one of the 27 conformations preferred by each amino acid residue. This still theoretically results in 27n conformations for a protein comprising ‘n’ amino acids. We then investigated the correlations at the 2-residue (dipeptide) and 3-residue (tripeptide) levels in what may be described as higher order Ramachandran maps, with the premise that the allowed conformational space starts to shrink as we add more and more residues. We found for instance, for a tripeptide which potentially can exist in any of the 273 ‘allowed’ conformations, two thirds of this space is redundant to 95% confidence level suggesting sequence context dependent preferred conformations. We then created a lookup table of preferred conformations at the dipeptide and tripeptide levels and correlated them with energetically allowed conformations. An alpha-numeric string and hence the tertiary structure can be generated for any sequence from the look-up table within a minute on a single processor and to a higher level of accuracy if this can be supplemented with a secondary structure predictor.